fast NIFTI operations in R
LC
1/17/2021
Intro
I collect here a few notes about carrying out fast operations (well let’s say as fast as possible…) on NIFTI images in R.
A huge inspiration and source of information was the Neuroimaging Analysis with R course by John Muschelli
NB: This is constantly a work in progress, and it will change from time to time.
Load NIFTI image
RNifti is the fastest library I found so far to load NIFTI files
library(RNifti)
library(dplyr)
library(microbenchmark)
# superfast load with RNifti::readNifti
bd <- sprintf("%s/data",getwd())
nii_file <- paste0(bd,"/icbm152_2009_brain.nii.gz")
nii <- RNifti::readNifti(nii_file)
Indexing
hist(nii[nii>0])
Flattening onto a vector
niivec <- c(nii)
Send idx/val of nnz voxels to a data.frame
df <- data.frame(
idx = which(nii > 0)
) %>%
mutate(val = nii[idx])
df %>% head() idx val
1 17039 20.91664
2 17040 20.86172
3 17041 20.56490
4 17042 20.86172
5 17043 20.91664
6 17235 21.50880
List of idx and mean val for different ranges of vals
rois <- list(
roi_1 = 60,
roi_2 = 71,
roi_3 = 90
)
lapply(rois, function(x) {
list(
roi_numba = x,
numba_voxels = which(nii > x) %>% length(),
meanvals = mean(nii[which(nii > x)])
)
}) %>% jsonlite::toJSON(pretty = T){
"roi_1": {
"roi_numba": [60],
"numba_voxels": [1412538],
"meanvals": [73.142]
},
"roi_2": {
"roi_numba": [71],
"numba_voxels": [767138],
"meanvals": [79.658]
},
"roi_3": {
"roi_numba": [90],
"numba_voxels": [353],
"meanvals": [90.4395]
}
}
View slices
With lapply %>% do.call
slices_numba = c(80,90,100)
imlist <- lapply(slices_numba, function(x) nii[ , ,x])
imstack <- do.call(rbind, imlist)
image(imstack)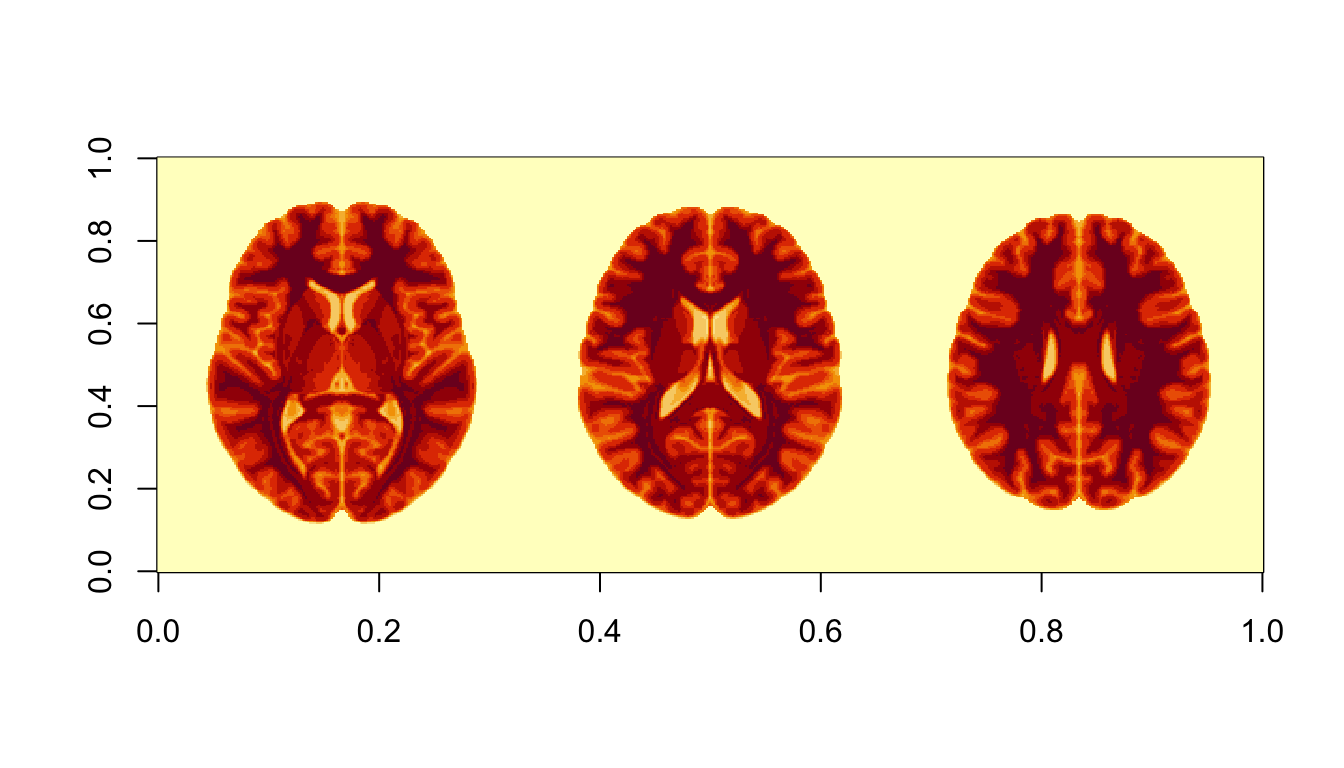
Manual slicing
sl80 <- nii[ , ,80]
sl90 <- nii[ , ,90]
sl100 <- nii[ , ,100]
image(rbind(sl80,sl90,sl100))
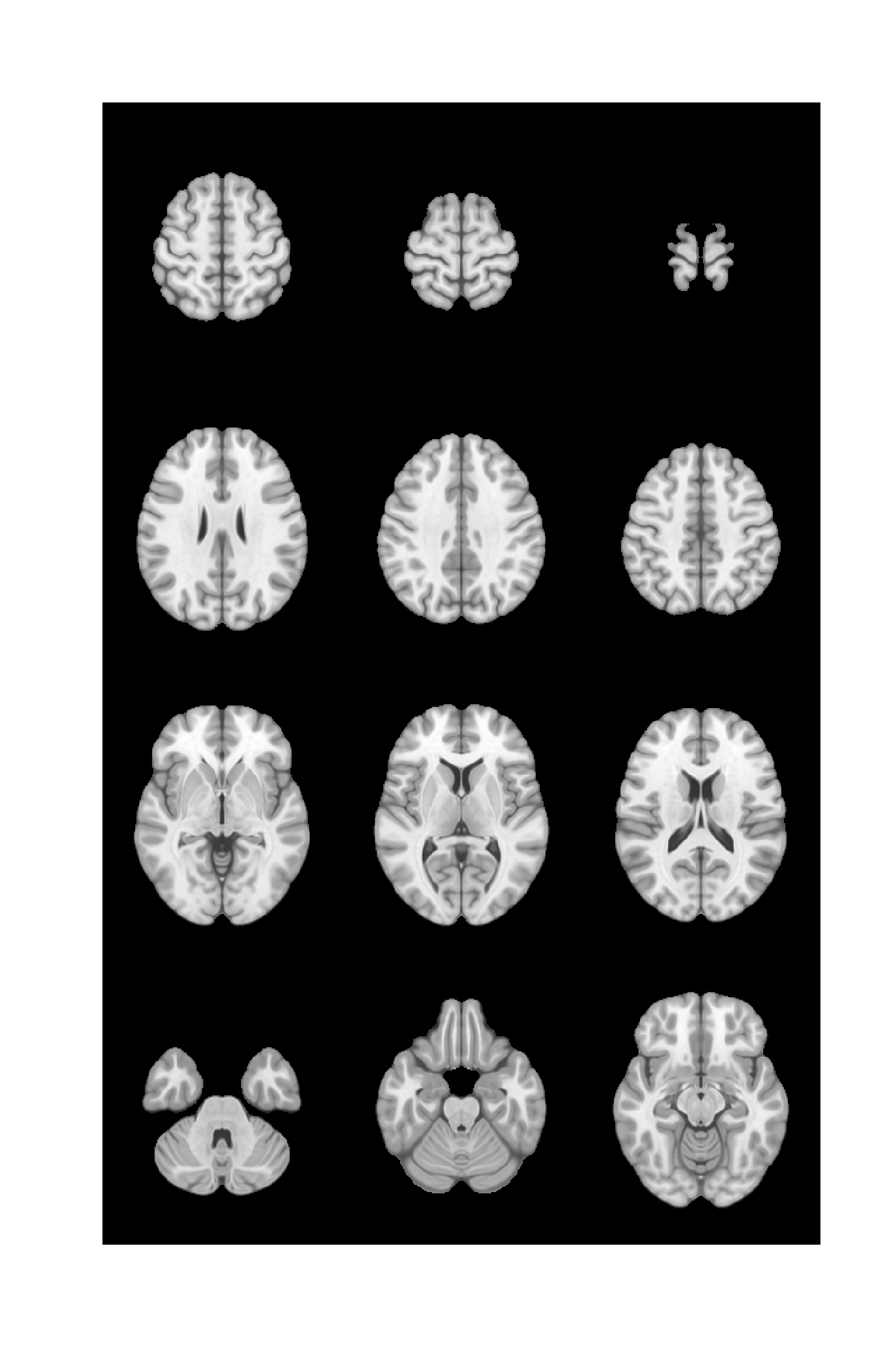
Build a 4x3 montage
library("RColorBrewer")
col <- colorRampPalette(c("black","white"))(256)
slices_numba = seq(40,170,10)[1:12]
imstack <- lapply(slices_numba, function(x) nii[ , ,x]) %>% do.call(rbind, .)
montage <- function(imstack,nr) {
ist <- imstack
# length of each row
xspan <- nrow(ist)/nr
# dummy to add rows
sq <- vector()
# add rows and delete them from ist
for (i in 1:nr) {
onerow <- ist[1:xspan, ] # select one row
sq <- cbind(sq,onerow) # add row to sq
ist <- ist[-c(1:xspan),] # delete row from ist
}
return(sq)
}
montage(imstack,nr=4) %>% image(col=col, axes=F, useRaster = T)